AI-Driven Digital Twin of Cell State Evolution and Perturbation
The advent of high-throughput omics technologies has enabled a more quantitative and integrative understanding of cellular dynamics. Our research focuses on developing AI-driven digital twins of cells, creating dynamic computational models that simulate cell state evolution and their perturbation in development, disease and therapeutic intervention. By integrating multi-omics data with deep learning, we aim to delucidate the transcriptional logic and regulatory networks that govern cell fate, moving from static observation to dynamic, predictive simulation of cellular processes and transitions.
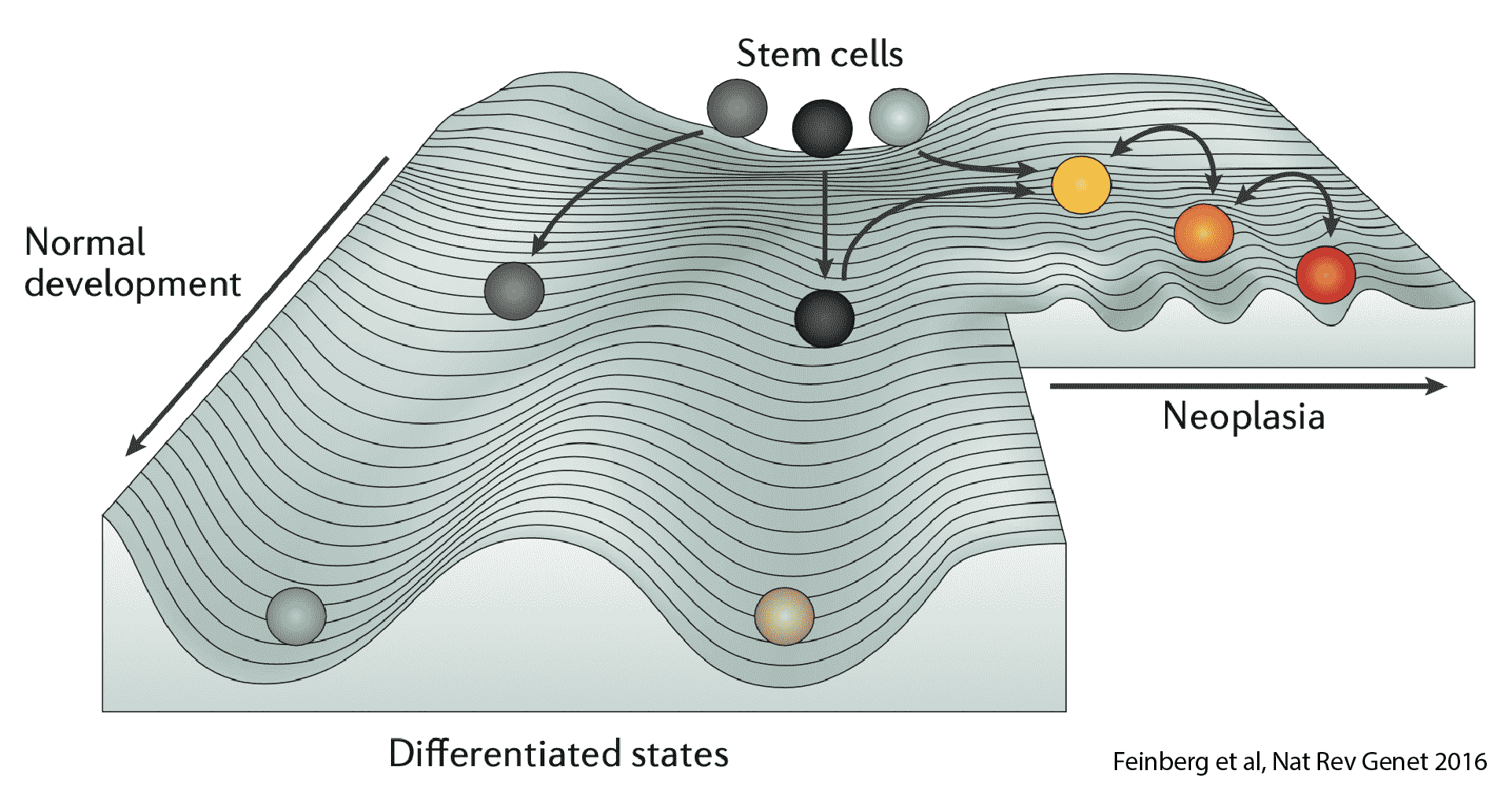
Major research directions:
- Integrate large-scale multi-omics data to develop deep learning models for cellular representation and state prediction.
- Establish a digital twin system for cells to simulate cellular dynamics and perturbation responses during development, homeostasis, disease progression, and therapeutic intervention.
- Elucidate the interactions among transcription factors and regulatory elements, and uncover the mechanisms by which cell-specific regulatory networks maintain states and facilitate transitions.
To achieve these goals, we use a wide range of cutting-edge technologies, including deep learning, bioinformatics, genomics, CRISPR gene editing, high-throughput screening, and next generation sequencing techniques (such as STARR-Seq、Hi-C、ATAC-Seq、ChIP-Seq、RNA-Seq and so on).